Scientific Program
Keynote Session:
Oral Session 1:
- Plant Genomic Science
Title: Genome-wide structural and functional landscapes of SNPs revealed from the WGRS data of 179 accessions of Arachis
Biography:
Ramesh S. Bhat has contributed significantly for the Post-Graduate teaching and Research in the area of Molecular Biology and Biotechnology. The major focus of his research has been the development of transposable element-based marker system, mapping of foliar disease resistance, taxonomic and drought tolerance traits, marker validation [using heterogenous inbred families and, various recombinant inbred line and multi-parent advanced generation inter cross (MAGIC) populations] and molecular breeding in peanut. Genomic location and properties of Arachis hypogaea miniature inverted repeat transposable element 1 (AhMITE1) were characterized. Genome-wide mutations were also studied in terms of transposon activity, SNP and CNV. Extensive analysis of DNA methylation and SNPs were carried out in peanut by his group. Two backcross lines with foliar disease resistance developed have been approved for commercial cultivation. Currently, the backcross lines developed in the background of two elite varieties (GPBD 4 and G2-52) are under field evaluation for variety development.
Abstract:
Peanut being an important food, oilseed and fodder crop worldwide, its genetic improvement currently relies on genomics-assisted breeding (GAB). Since the level of marker polymorphism is limited in peanut, availability of a large number of DNA markers is the prerequisite for GAB. Therefore, we detected 4,309,724 single nucleotide polymorphisms (SNPs) from the whole genome re-sequencing (WGRS) data of 178 peanut accessions of Arachis along with the reference genome sequence of Tifrunner. SNPs were analyzed for the structural and functional features in order to conclude on their utility and employability in genetic and genomic studies. ISATGR278-18, a synthetic amphidiploid, showed the highest number of SNPs (2,505,266), while PI_628538 recorded the lowest number (19,058) of SNPs. A03 showed the highest number of SNPs, while the B08 recorded the lowest number of SNPs. The number of accessions required to record 50% of the total SNPs varied from 11 to 13 across the chromosomes. The rate of transitions was more than that of transversions. Among the various chromosomal contexts, intergenic and intronic regions carried more SNPs than the exonic regions. SNP impact analysis indicated 2,488 SNPs with high impact due to gain of stop codons, variations in splice acceptors and splice donors, and loss of start codons. Of the 4,309,723 SNPs, as high as 46,087 had the highest polymorphic information content (PIC) of 0.375. As an illustration of application, a drought and disease resistant accessions were compared with susceptible accessions to identify the SNPs with high impact substitutions. SNPs were also identified between the two sub-genomes (A and B). This SNP resource is being used for haplotype selection and allele mining for various genes and gene families. Overall, these structural and functional landscapes of the SNPs will be of immense importance for their utility in genetic and genomic studies including reverse genetics in peanut.
Title: Nano-assisted Genome Editing for Crop Improvement
Biography:
Shailendra Singh Gaurav, Dean, Faculty of Agriculture, Professor & Ex-Head, Department of Genetics & Plant Breeding, Ch. Charan Singh University, Meerut (UP) India has joined this department as Lecturer in December 2001 after completion of his doctoral degree from ICAR- Indian Agricultural Research Institute, Pusa, New Delhi. Currently, he is also Head departments of biotechnology, Seed Science & Technology, Intellectual property Cell, All India Survey on Higher Education Cell as coordinator/nodal officer of this university. Prof. Gaurav is presently engaged in the research activities are focused on the different areas of Plant Breeding, Seed technology and plant biotechnology to address the local concern of Western UP that includes morphological , biochemical & molecular characterization and evaluation of genetic diversity, heterosis breeding in various crops like wheat, rice, lemon grass, basil, chickpea, pea etc. Recently, Prof. Gaurav & his team have initiated their research work to exploit the possibilities for the development of nano-bio pesticides against early & late blight of potato and white grub of sugar cane crop. Dr. Gaurav is also engaged in teaching of Masters (M.Sc. & M.Phil) and Doctoral programme. His research contributions, he has been able to publish 99 original research papers in peer-reviewed journals of national and international repute and 01 patent credited to him. Apart from this, he has also authored 06 text books, 10 popular articles, 04 book chapters.
Abstract:
Current agricultural practices need to be modernized and strengthened to meet the food needs of the growing world population. Hence, new and advanced approaches keep on developing for the crop-improvement. Nano-enabled agriculture is a new such area. Genome editing using nanotechnology is used in agricultural research to improve stress tolerance, targeting, controlled release of pesticides, and improve photosynthetic efficiency. Genome editing is more accurate, faster, and cheaper than other transgenic approaches. Clustered regularly interspaced short palindromic repeats (CRISPRs) using nucleases are a customizable and successful method for genome editing. However, there are some limitations such as time-consuming and complex protocols, inevitable DNA integration into the host genome, potential tissue damage, and low transformation efficiency. To overcome these problems, nanoparticle-mediated gene delivery has emerged as an innovative technology. Combining current technologies such as speed breeding and CRISPR/Cas with nanotechnology can advance crop improvement programs. Recent advances in the development of genome editing tools have revolutionized the ability of researchers to genetically investigate and modify biological systems. However, genetic engineering of mature plants and their plastids remains a challenge due to the various physical barriers. Nanomaterials develop a nanoscale understanding of the mechanisms that can be used to achieve nanoparticle transport across plant cells and chloroplast membranes and identify highly efficient nanoparticle complexes for plant cell internalization. Researchers are creating a nanoparticle platform that enables electrostatic-grafting of genomic biomolecules used to genetically transform mature plants. In addition to the use of magnetic nanoparticles, carbon nanotubes are also an increasingly popular nano-tool for plant transfection. Carbon nanotubes merged with gene fragments have been shown to effectively transport transient and expression genes to the leaves of various plant species, including wheat, cotton, and arugula. Nanotechnology-based genomics is still in its infancy, but it has a promising future for crop improvement and modern agricultural research.
Oral Session 2:
- Plant Molecular Biology
Title: The molecular mechanism behind the primed seed of cotton (Gossypium hirsutum L.)
Biography:
Ghulam Abbas has his expertise in seed priming, soil salinity (Plant Stress Physiology), and passion for improving the health of saline soil. He is working on different methods to find out suitable techniques to grow crops in saline soil. He is also working on molecular tools to find out and improve genes responsive to salt tolerance, particularly in cotton. He is also currently working usefulness of Algal extract (bio-priming) against the environmental stress of different crop plants. He did work on physical, physiological, and molecular aspects of seed priming under salinity stress during his Masters’ project. His work to develop a sustainable approach that will offer a new area of farming for farmers and the economy.
Abstract:
The molecular mechanisms behind the seed priming are still a mystery. However, advanced molecular tools such as genomics, transcriptomics, and proteomics have added significant information. Seed priming is a seed pre-soaking technique that is used to initiate pre-germination processes inside by activating the expression of certain genes. This study was designed to reveal important molecular mechanisms behind cotton seed priming. Seed priming completes into three phases: I. Imbibition phase, II. Lag phase, and III. Radicle emergence phase. Proteomic analysis revealed that GhAKT1 (K+ channel gene) was activated and expressed during phase I of primed seeds and facilitated the transport of K+ and elongation of embryonic cells in primed seeds. The repair and synthesis of new mitochondria were also observed in primed seeds by the resumption of respiratory activities. However, expression of ACT7 (Actin-gene) was also observed in primed seeds. In Phase II, more RNA transcripts were found than unprimed seeds. Several events were revealed, including the mitochondrial repair, synthesis of new mitochondria, and proteins. The embryonic expansion also began due to the expression of the GhAKT1 gene. The proteomic analysis also revealed the synthesis of α-amylase, β-amylase, protease, and isocitrate lyase in primed seeds which are required for degradation and translocation of reserve food. The end of Phase II and the start of Phase III marked by radicle protrusion through the testa/seed coat. During the third phase, there was a significant increase of actin proteins found in primed seeds which characterized cell division. Radicle cells elongation was also noted by following the activity of ACT7, and GhAKT1 genes.
Title: Genome-wide investigation of 20S proteasome family genes and their relevance to heat and drought tolerance in common wheat
Biography:
Sachin Kumar is Assistant Professor of Genetics and Plant Breeding at Chaudhary Charan Singh University, Meerut, India. His research area covers Plant Genomics and Molecular Breeding in cereal crops particularly wheat, which include trait discovery and phenotyping, high-throughput SNP genotyping, construction of high-density genetic maps, physical and radiation hybrid maps, comparative mapping, QTL interval mapping, genome-wide association studies (GWAS), development of molecular markers, marker-assisted selection for biotic and abiotic stress-responsive traits. He has been NSERC Visiting Fellow under Canadian Government Laboratories Program at Agriculture and Agri-food Canada (AAFC), Swift Current, Saskatchewan, Canada.
Abstract:
The genes encoding α- and β-type subunits of 20S proteasome core protease are integral components of 26S proteasome complex in the ubiquitin-proteasome system (UPS). The 20S proteasome represents a catalytic particle that cleaves cytotoxic, denatured, damaged and unwanted proteins in an ATP/ubiquitin-dependent non-lysosomal pathway and is known to impart thermotolerance in model plants. In the present study, we identified a family of 67 genes (including 34 TaPA and 33 TaPB genes) encoding α- and β-subunits of 20S proteasome in bread wheat (Triticum aestivum L.). These genes were distributed on all the 21 wheat chromosomes, majority of them being in triplicate homoeologues. These wheat genes were orthologous to corresponding rice and Arabidopsis genes. The proteins encoded by each of the TaPA and TaPB genes contained 20 different motifs. These 20 motifs were present in corresponding proteins of Arabidopsis and rice. Phylogenetic analysis placed these genes in seven clusters, each with one of the seven α (α1-7) and one of the seven β (β1-7) subunits. Expression analysis suggested that 10 of the 67 genes were involved in heat stress response, whereas four genes were involved in drought tolerance at the seedling stage. Nine (9) genes were expressed under both heat and drought suggesting their involvement in response to multiple abiotic stresses. Future research on TaPA/TaPB genes will help in the development of climate-resilient wheat cultivars.
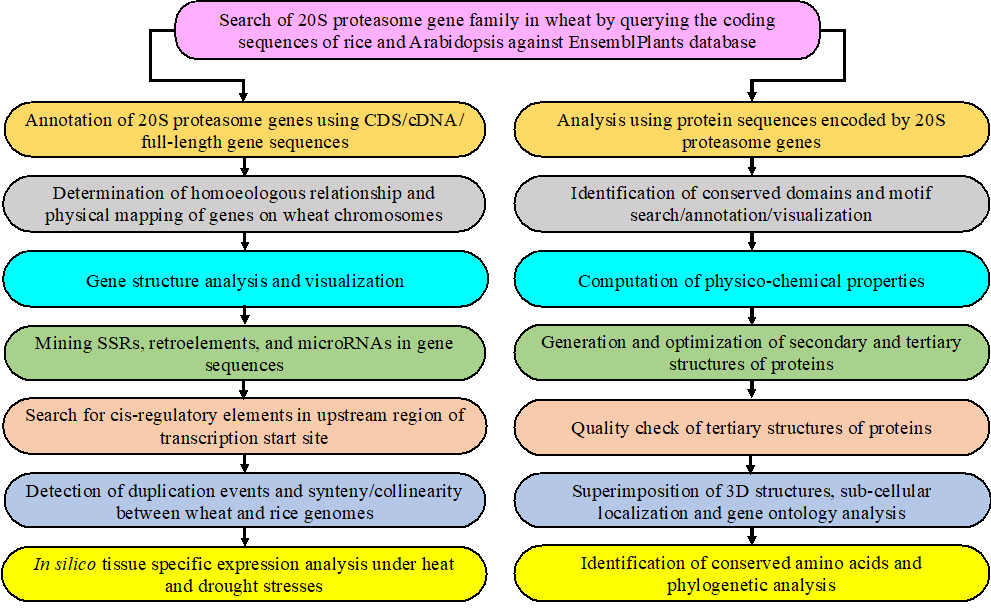
FIG. Schematic representation of the steps and programs used to identify 20S proteasome family genes in bread wheat.





